Techniques
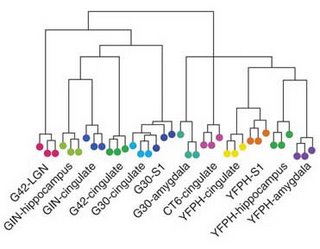
like this on a larger-scale might help get a handle on nervous system evolution. They took several mouse lines expressing fluorescent proteins under the control of different promoters and isolated individual cells from brain regions like the cingulate cortex, somatosensory cortex, hippocampus, and amygdala. They then used a microarray to look at the level of ~13,000 mRNAs within each cell type and measured the distance between the expression profiles to generate a tree. The major distinction between excitatory and inhibitory neurons (glutamatergic vs. GABAergic) was the first split. Cortical pyramidal neurons (the bulk of cortical neurons) were the most closely related by this measure. There are pyramidal neurons in the hippocampus as well and these weren't too far from the cortical version. Interesting to see how closely the neocortex stuck to the template of allocortex cell-types as it evolved. Lots of interesting tidbits. The genes that are expressed at different levels in different cell-types tend to be part of gene families. The authors claim this provides support for the notion of gene duplication loosening the clamp of selection a little bit. I've been curious about how much one can generalize experimental results from hippocampal to neocortical pyramidal cells and vice versa. I'll tell you why if I ever find time to discuss
these papers.
Neuronal cell-type dendrogram below the fold: